The Emperor of All Maladies’ New Clothes
April 8, 2015 1 Comment
Ken Burn’s series “The Emperor of All Maladies” from Siddhartha Mukherjee’s book of the same title p rovides an interesting and informative historical perspective on mankind’s efforts to confront cancer as a disease.
rovides an interesting and informative historical perspective on mankind’s efforts to confront cancer as a disease.
Beginning with ancient references to human malignancy, the series goes on to explore radical surgery and the earliest use of radiation but really gains traction in the mid-20th century with the discovery of the first chemotherapy drugs. While the nitrogen mustard derivatives were being studied under a veil of military secrecy, Dr. Sidney Farber in Boston explored the B-vitamin analogue, aminopterin, for the treatment of childhood leukemia. (You can read more about this in my book Outliving Cancer.)
Through the ensuing decades, seemingly stunning victories ultimately fell in crushing defeats, while the promise of single agents, then multi-drug combinations, followed by dose-intensive therapies, and finally bone marrow transplantation yielded few cures but delivered ever increasing toxicities. Clifton Leaf, a cancer survivor himself who created a stir with his controversial 2003 Fortune Magazine article entitled “Why We Are Losing the War on Cancer and How to Win It” described his own disappointment with the slow pace of progress.
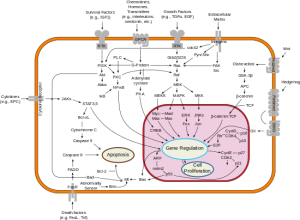
 The last episode examined our growing understanding of human genomics and segued by interviews with Richard Klausner, former director of the National Cancer Institute; and Harold Varmus, the current NCI director; to Michael Bishop, Eric Lander and Francis Collins who luxuriated in the clinical potential of human genomics and the coming era of big science.
The last episode examined our growing understanding of human genomics and segued by interviews with Richard Klausner, former director of the National Cancer Institute; and Harold Varmus, the current NCI director; to Michael Bishop, Eric Lander and Francis Collins who luxuriated in the clinical potential of human genomics and the coming era of big science.
The final part was an interview with Steven Rosenberg, one of the earliest pioneers in immunotherapy and Carl June whose groundbreaking work with chimeric antigen receptor T-cells is among the most recent applications of this important field.
The take-home message would seem to be that despite the fits and starts we are now at the dawn of a new age of big science, big data and genomic breakthroughs. What was missing however was an examination of where we had gone wrong. It would seem that the third rail for this community is an honest assessment of how a small coterie of investigators who championed only certain ways of thinking over all others commandeered all the money, grants, publications, chairmanships and public attention, while patients were left to confront a disease from which survival has changed very little, at ever increasing costs and toxicities.
Another thing that came through was the very human side of cancer as a disease and the kindness and emotional support that family members and parents provided to those afflicted. I couldn’t help but feel that these individuals had been cheated: cheated of the lives of their family members, cheated of the resources that could have pursued other options and cheated of the well-being that these poisonous and dose-intensive regimens rained upon them in their last days.
As science has become the new religion and scientists the new gurus, one message that resonated was that many of these gurus were false prophets. They are too self-absorbed to question their own dogmatic belief systems in dose-intensity or multi-agent combinations, all of which fell painfully by the way side as the next therapeutic fad emerged. Will our current love affair with the gene prove to be little more than the most current example of self-congratulatory science conducted in the echo chamber of modern academia?
Victories against cancer will be won incrementally. Each patient must be addressed as an individual, unique in their biology and unique in their response probability. No gene profile, heat map, DNA sequence or transcriptomic profile has answered the questions that every patient asks; “What treatment is best for me?” Dr. Mukherjee himself used the analogy of the blind men and the elephant. Unfortunately, there was little discussion of how much that parable may apply to our current scientific paradigms.
It is time for patients to demand better and refuse to participate in cookie-cutter protocols.

Physicians should become more familiar with the fundamentals of physiology and biochemistry to better understand the principles of cancer prevention at the level of diet and lifestyle.
Finally, while we wait with bated breath, for the arrival of glorious gene profiles widely touted as the future answer to all of cancer’s most vexing questions, patients should throw off the yoke of one-size-fits-all approaches and demand laboratory platforms, such as the EVA-PCD assay, that are available today to make better use of existing treatments.