What is Personalized Cancer Therapy?
June 24, 2015 Leave a comment
Personalized therapy is the right treatment, at the right dose for the right patient. Like the weather, however, it seems that everyone’s talking about it, but no one is doing anything about it.
In its simplest form personalized care is treatment that is designed to meet an individual’s unique biological features. Like a key in a lock, the right drug or combination opens the door to a good outcome.
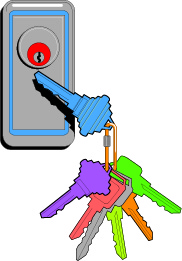 When over the years I lectured on the development of the cisplatin/gemcitabine doublet, my two boys were quite young. I would show a slide depicting a doorknob with a key in the keyhole. I likened our lab’s capacity to identify sensitivity to the cisplatin/gemcitabine combination as “unlocking” an individual’s response.
When over the years I lectured on the development of the cisplatin/gemcitabine doublet, my two boys were quite young. I would show a slide depicting a doorknob with a key in the keyhole. I likened our lab’s capacity to identify sensitivity to the cisplatin/gemcitabine combination as “unlocking” an individual’s response.
At the time my wife and I would leave the key in the inside of the front door enabling us to unlock it when going out. We reasoned at the time that our 2-year-old would not be strong enough, nor tall enough to turn the key and let himself outside. We reasoned wrong, for one day our son Alex reached up, turned the key and opened the door right in front of us. Lesson learned: Given the right key, anyone can open a door.
I continued my analogy by saying that even Arnold Schwarzenegger would be unable to open a door given the wrong key, but might, if he continued trying, snap it off in the lock.
The right key is the right treatment, effortlessly unlocking a good response, while the wrong key is the wrong treatment more often than not too much, too late, akin to a solid tumor bone marrow transplant.
In recent years, personalized care has come to be considered synonymous with genomic profiling. While we applaud breakthroughs in human genomics today, there is no molecular platform that can match patients to treatments. The objective response rate of just 10 percent, almost all in breast and ovarian cancer patients in one study (Von Hoff J Clin Oncol 2010 Nov 20:28(33): 4877-83), suggests that cancer biology is demonstrably more complex than an enumeration of its constituent DNA base pairs. The unilateral focus on this area of investigation over others might be described as “the triumph of hope over experience” (James Boswell, Life of Samuel Johnson, 1791).
But hope springs eternal and with it the very real possibility of improving our patients outcomes. By accepting, even embracing, the complexity of human tumor biology we are at the crossroads of a new future in cancer medicine.
William Withering (1741-1799) the English physician and botanist credited with discovering digitalis as the therapy for dropsy, e.g. congestive heart failure (An Account of the Foxglove and some of its Medical Uses, Withering W. 1785), had absolutely no idea what a membrane ATPase was, when he made his remarkable discovery. It didn’t matter. Cardiac glycosides provided lifesaving relief to those who suffered from this malady for fully two centuries before Danish scientist, Jens Christian Skou, identified these membrane bound enzymes, for which he was awarded a Nobel Prize in 1997.
Similarly, penicillin, aspirin, and morphine were in all use for decades, centuries, even millenia before their actual modes of action were unraveled. Medical doctors must use any and all resources at their disposal to meet the needs of their patients. They do not need to know “how” something works so much as they (and their patients) need to know “that” it works.
The guiding principle of personalized medicine is to match patients to therapies. Nowhere in this directive is there a prescription of the specific platform to be used. Where genomic signatures provide useful insights for drug selection, as they do in APL (ATRA, Arsenic trioxide); NSCLC (EGFr, ROS1, ALK); CML (Imatinib, Dasatanib) then they should be used.
However, in those disease where we haven’t the luxury of known targets or established pathways, i.e. most human malignancies, then more global assessments of human tumor biology should, indeed must, be used if we are to meet the needs of our patients. Primary culture analyses like the EVA-PCD® provide a window onto human tumor biology. They are vehicles for therapy improvement and conduits for drug discovery. Scientists and clinicians alike need to apply any and all available methodologies to advance their art. The dawn of personalized medicine will indeed be bright if we use all the arrows in our quiver to advance clinical therapeutics and basic research.
Reposted from May 2012